Técnicas de IA para Biologia
6 - Representation Learning
André Lamúrias
Representation Learning
Motivation
Motivation
Features are very important
- Example: long division using Arabic or Roman numerals
- In machine learning, the right representation makes all the difference
- Deep learning can be seen as stacked feature extractors, until the final classification
- The top layer could even be replaced by another type of model, in theory
Representation learning
- Supervised learning with limited data can lead to overfitting
- But learning the best representation can be done with unlabelled data
- Unsupervised and semi-supervised learning can help find the right features
Motivation
"Meta-priors": what makes a good representation?
Representation Learning; Bengio, Courville, Vincent, 2013
- Manifolds:
- Actual data is distributed in a subspace of all possible feature value combinations
- Disentanglement:
- Data is generated by combination of independent factors (e.g. shape, color, lighting, ...)
- Hierarchical organization of explanatory factors:
- Concepts that explain reality can be composed of more elementar concepts (e.g. edges, shapes, patterns)
- Semi-supervised learning:
- Unlabelled data is more numerous and can be used to learn structure
Motivation
"Meta-priors": what makes a good representation?
Representation Learning; Bengio, Courville, Vincent, 2013
- Shared factors:
- Important features for one problem may also be important for other problems (e.g. image recognition)
- Sparsity:
- Each example may contain only some of the relevant factors (ears, tail, legs, wings, feet)
- Smoothness:
- The function we are learning outputs similar $y$ for similar $x$
Motivation
"Meta-priors": what makes a good representation?
Representation Learning; Bengio, Courville, Vincent, 2013
- If we can capture these regularities, we can extract useful features from our data
- These features can be reused in different problems, even with different data
Representation Learning
Unsupervised Pretraining
Unsupervised Pretraining
Greedy layer-wise unsupervised pretraining
- Greedy: optimizes each part independently
- Layer-wise: pretraining is done one layer at a time
- E.g. train autoencoder, discard decoder, use encoding as input for next layer (another autoencoder)
- Unsupervised: each layer is trained without supervision (e.g. autoencoder)
- Pretraining: the goal is to initialize the network
- It is followed by fine-tuning with backpropagation
- Or by training of a classifier "on top" of the pretrained layers
Unsupervised Pretraining
Why should this work?
- Initialization has regularizing effect
- Initially thought as a way to find different local minima, but this does not seem to be the case (ANN do not generally stop at minima)
- It may be that pretraining allows the network to reach a different region of the parameter space
Unsupervised Pretraining
When does it work?
- Poor initial representations
- E.g. word embedding from one-hot vectors
- One-hot vectors are all equidistant, which is bad for learning
- Unsupervised pretraining helps find representations that are more useful
- Example: (Large) Language Models
- Known as self-supervised learning, latent representations are called word embeddings
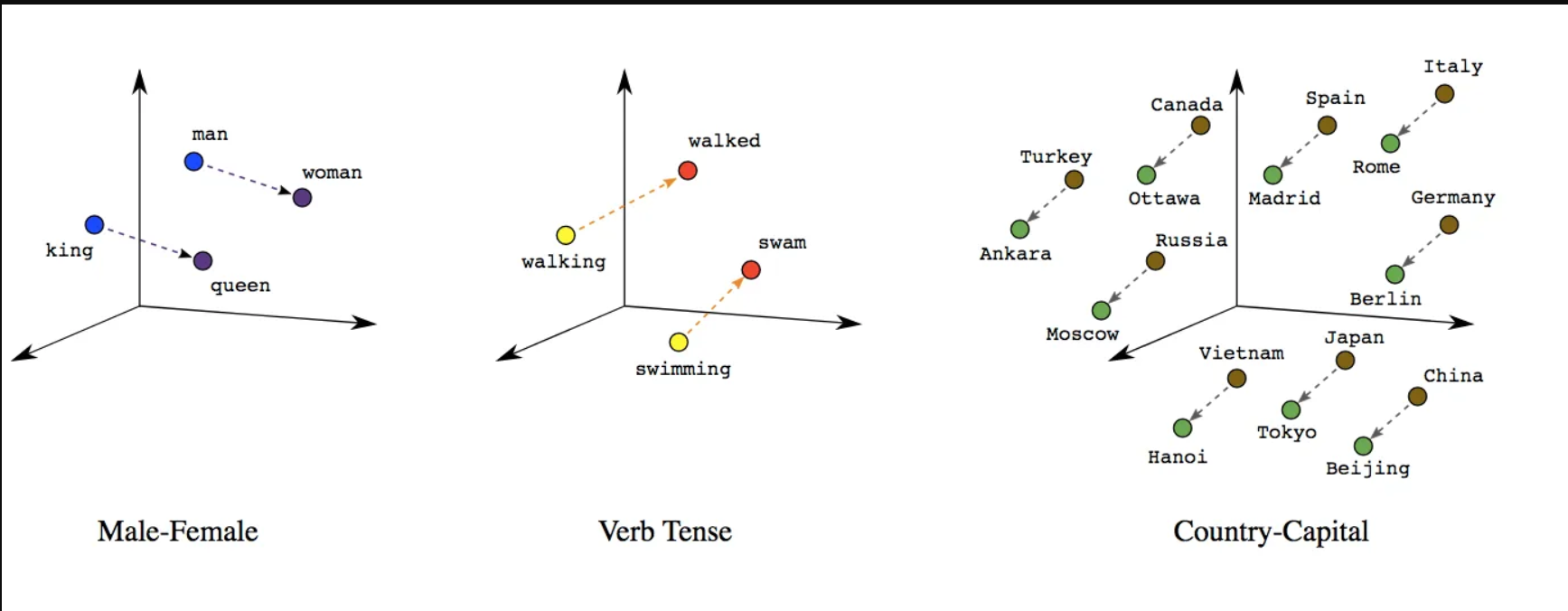
https://developers.google.com/machine-learning/crash-course/embeddings/translating-to-a-lower-dimensional-space
Unsupervised Pretraining
When does it work?
- Poor initial representations
- E.g. word embedding from one-hot vectors
- One-hot vectors are all equidistant, which is bad for learning
- Unsupervised pretraining helps find representations that are more useful
- Regularization, for few labelled examples
- If labelled data is scarce, there is greater need for regularization and unsupervised pretraining can use unlabelled data for this
- Example: Training trajectories with and without pretraining
- Concatenate vector of outputs for all test set at different iterations
- (50 nertworks with and without pretraining)
- Project into 2D (tSNE and ISOMAP)
Unsupervised Pretraining
- Regularizing effect
- Ehrlan et. al. 2010: output vectors for all data, reduce dimensionality, plot

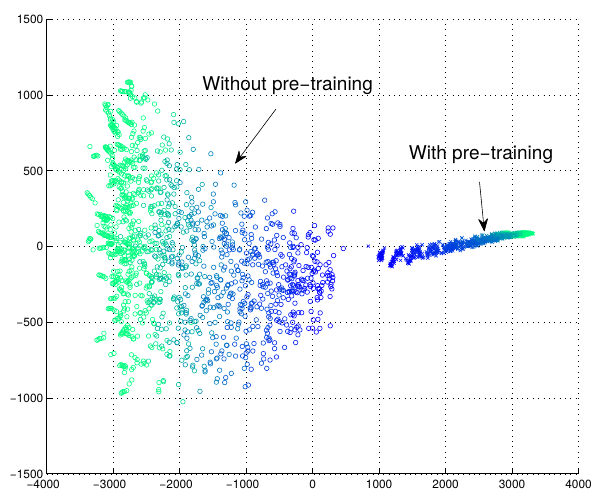
t-Distributed Stochastic Neighbor Embedding and ISOMAP
Unsupervised Pretraining
Unsupervised pretraining is historically important
- It was the first practical method for training deep networks
- Most DL problems habed abandoned it because of ReLU and dropout, which allows efficient supervised training and regularization of the whole network
- For very small datasets, other methods outperform neural networks
- e.g. Bayesian methods
- Another disadvantage: having two training stages makes it harder to adjust Hyperparameters
- Commonly used in some applications, such as natural language processing
- Unsupervised pretraining with billions of examples to learn good word representations
Representation Learning
Transfer Learning
Transfer Learning
Two different tasks with shared relevant factors
- Shared lower level features:
- E.g. distinguish between cats and dogs, or between horses and donkeys
- The low level features are mostly the same, only the higher level classification layers need to change
- Shared higher level representations:
- E.g. speech recognition
- The high level generation of sentences is the same for different speakers
- However, the low level feature extraction may need to be tailored to each speaker
Domain adaptation
Same underlying function but different domains
- We want to model the same mapping from input to output, but are using different sets of examples
- E.g. sentiment analysis
- Model was trained on customer reviews for movies and songs
- Now we need to do the same for electronics
- There should be only one mapping from words to happy or unhappy, but we are training on different sets with different words
- This is one example where unsupervised training (DAE - denoising autoencoders) can help
Concept Drift
Similar to Transfer learning or domain adaptation
- But occurs when the change is gradual over time
- E.g. as the brand becomes more popular, customer base changes from specialized to general
Transfer Learning
Use previous experience in new conditions
- The common idea is that we can use what was learned before to help learn now
- Extreme examples: one-shot learning and zero-shot learning
- Zero-shot learning: no labelled examples of new classes are necessary
- Everything was learned on other classes or unsupervised
- One-shot learning: use only one labelled example to learn new dataset
- The rest was learned on other data
Transfer Learning
- Zero-shot learning, example:
- Unsupervised learning of word manifold, supervised mapping of known images
- A new image is mapped to word manifold
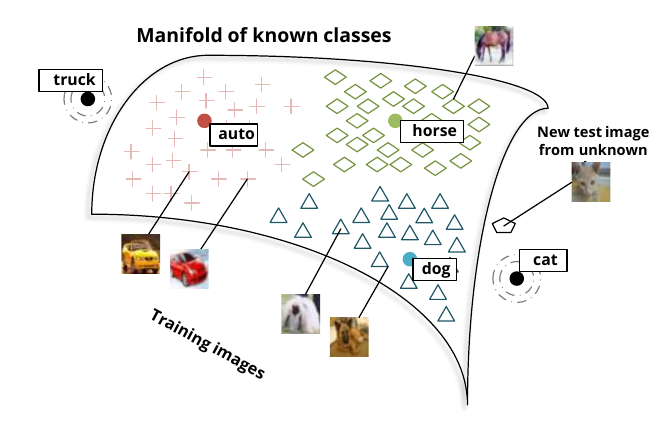
Socher et. al. Zero-Shot Learning Through Cross-Modal Transfer (2013)
Representation learning
Summary
Representation learning
Summary
- Improve learning from poor representations
- Find the best features
- Regularization or feature extraction with unlabelled data
- Historically important in deep learning
- Still used in NLP (self-supervised learning)
- Transfer learning (often supervised)
Further reading
- Goodfellow et.al, Deep learning, Chapter 15 (and 8.7.4)
- Bengio et. al. Representation Learning: A Review and New Perspectives, 2013
Representation Learning
Exercise 1: transfer learning
Fashion MNIST to MNIST
Exercise
Transfer learning
- Use network trained on Fashion MNIST
Exercise
Transfer learning
- Use network trained on Fashion MNIST


- Use Keras functional API and pre-trained model
Exercise
Transfer learning
- Using pre-trained model:
- Create same graph as model we trained last week
- Load weights from last week's trained model
- Fix weights of convolutional part (won't be retrained)
- Recreate the dense classifier and train it on MNIST
Exercise
- Prepare dataset
- Same process as last week, but with MNIST instead of Fashion MNIST
from tensorflow import keras
from tensorflow.keras.optimizers import SGD
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Input, BatchNormalization, Conv2D, Dense
from tensorflow.keras.layers import MaxPooling2D, Activation, Flatten, Dropout
((trainX, trainY), (testX, testY)) = keras.datasets.mnist.load_data()
trainX = trainX.reshape((trainX.shape[0], 28, 28, 1))
testX = testX.reshape((testX.shape[0], 28, 28, 1))
trainX = trainX.astype("float32") / 255.0
testX = testX.astype("float32") / 255.0
Exercise
- Keras functional API
- Layer objects are callable (implement
__call__method) - Receive tensors as arguments, return tensors
-
Inputinstantiates a Keras tensor (shaped like the data) - Naming layers helps finding them again in the model
- We start by recreating the original model architecture
inputs = Input(shape=(28,28,1),name='inputs')
layer = Conv2D(32, (3, 3), padding="same", input_shape=(28,28,1))(inputs)
layer = Activation("relu")(layer)
layer = BatchNormalization(axis=-1)(layer)
layer = Conv2D(32, (3, 3), padding="same")(layer)
layer = Activation("relu")(layer)
layer = BatchNormalization(axis=-1)(layer)
layer = MaxPooling2D(pool_size=(2, 2))(layer)
layer = Dropout(0.25)(layer)
...
Exercise
- Chain layers as in the original model
- Create the old model and compile it
- Load previous weights and disable training in the old model
- Note:
Flattenlayer named for use in new model
...
features = Flatten(name='features')(layer)
layer = Dense(512)(features)
layer = Activation("relu")(layer)
layer = BatchNormalization()(layer)
layer = Dropout(0.5)(layer)
layer = Dense(10)(layer)
layer = Activation("softmax")(layer)
old_model = Model(inputs = inputs, outputs = layer)
old_model.compile(optimizer=SGD(), loss='mse')
old_model.load_weights('fashion_model')
for layer in old_model.layers:
layer.trainable = False
Exercise
- Create new dense layers for new model
- The input for this layer is the
"features"layer in old model - Then create the new model with:
- Old model
inputsas input - New softmax layer as output
- This chains old CNN to new dense layer
layer = Dense(512)(old_model.get_layer('features').output)
layer = Activation("relu")(layer)
layer = BatchNormalization()(layer)
layer = Dropout(0.5)(layer)
layer = Dense(10)(layer)
layer = Activation("softmax")(layer)
model = Model(inputs = old_model.get_layer('inputs').output, outputs = layer)
Exercise
- Now train
NUM_EPOCHS = 5
BS=512
opt = SGD(lr=1e-2, momentum=0.9)
model.compile(loss="categorical_crossentropy",
optimizer=opt, metrics=["accuracy"])
model.summary()
H = model.fit(trainX, trainY,validation_data=(testX, testY),
batch_size=BS, epochs=NUM_EPOCHS)
- Summary:
Total params: 1,679,082 Trainable params: 1,612,298 Non-trainable params: 66,784
- Compare with training all model from the start
Representation learning
Exercise 2: dimension reduction with autoencoder
Autoencoder
- Project MNIST data set into 2D

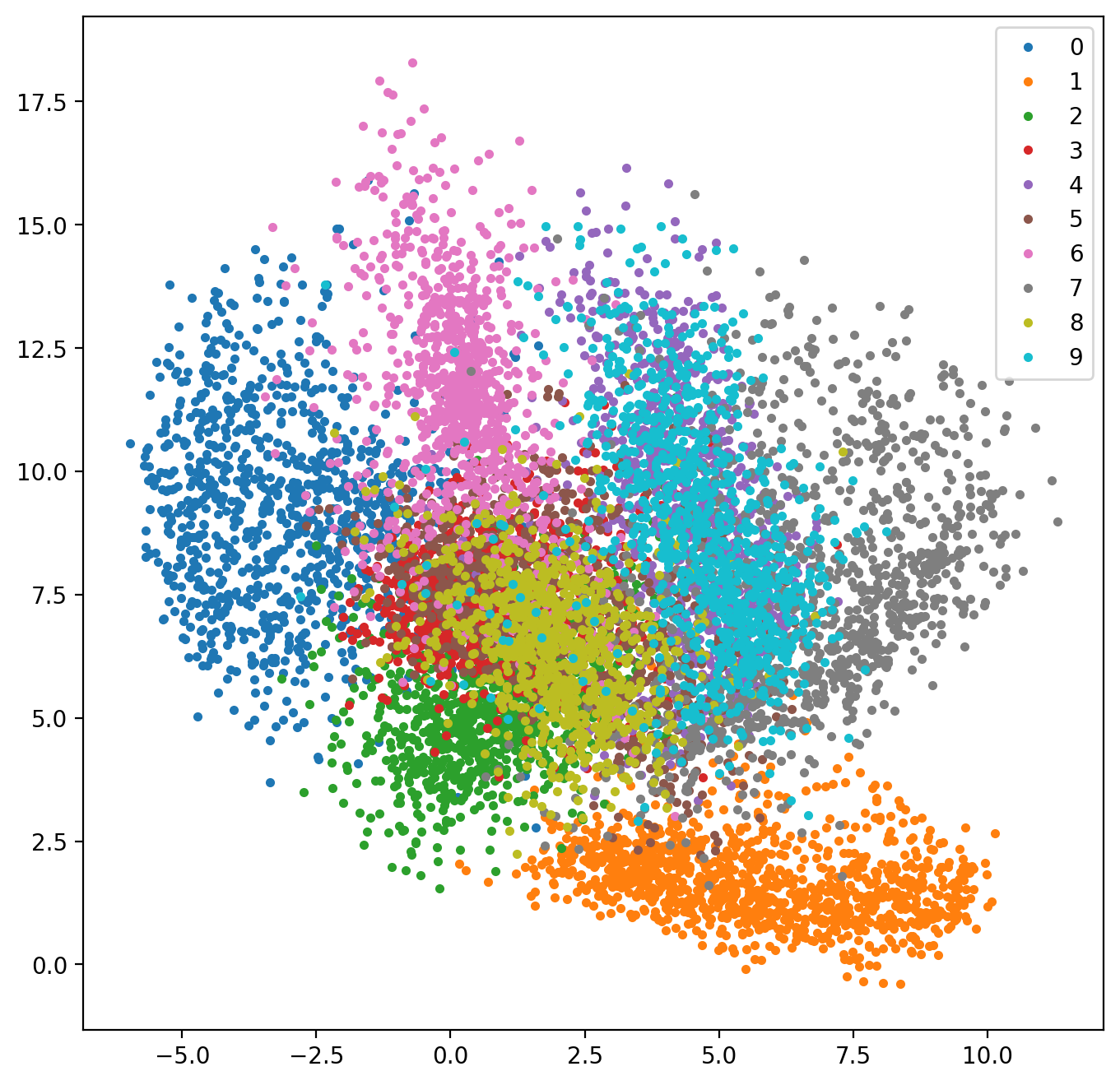
Autoencoder
- Need some extra layers:
from tensorflow.keras.layers import UpSampling2D,Reshape
- Encoder:
def autoencoder():
inputs = Input(shape=(28,28,1),name='inputs')
layer = Conv2D(32, (3, 3), padding="same", input_shape=(28,28,1))(inputs)
layer = Activation("relu")(layer)
layer = BatchNormalization(axis=-1)(layer)
#[...] parts missing here!
layer = MaxPooling2D(pool_size=(2, 2))(layer)
#[...] parts missing here!
layer = Conv2D(8, (3, 3), padding="same")(layer)
layer = Activation("relu")(layer)
layer = BatchNormalization(axis=-1)(layer)
layer = Flatten()(layer)
features = Dense(2,name='features')(layer)
Autoencoder
- Decoder:
layer = BatchNormalization()(features)
layer = Dense(8*7*7,activation="relu")(features)
layer = Reshape((7,7,8))(layer)
layer = Conv2D(8, (3, 3), padding="same")(layer)
layer = Activation("relu")(layer)
layer = BatchNormalization(axis=-1)(layer)
layer = Conv2D(16, (3, 3), padding="same")(layer)
layer = Activation("relu")(layer)
layer = BatchNormalization(axis=-1)(layer)
layer = UpSampling2D(size=(2,2))(layer)
[...]
layer = Conv2D(32, (3, 3), padding="same")(layer)
layer = Activation("relu")(layer)
layer = BatchNormalization(axis=-1)(layer)
layer = Conv2D(1, (3, 3), padding="same")(layer)
layer = Activation("sigmoid")(layer)
autoencoder = Model(inputs = inputs, outputs = layer)
encoder = Model(inputs=inputs,outputs=features)
return autoencoder,encoder
Autoencoder
Suggestions
- Architecture:
- Convolution layer with 32 filters, followed by pooling
- Convolution layer with 32 filters, then convolution layer with 16 filters, then pooling
- Convolution layer with 16 filters, then convolution layer with 8 filters, then pooling
- Dense layer with 2 neurons and linear output, then dense layer with 8*7*7 neurons (relu) and reshape
- Convolutions in reverse (8, 16, 16, 32 and 32 filters), and upsampling
- A final convolution of 1 filter for the (sigmoid) output.
- Training: takes some time (40 epochs or more...)
- Details weigh little on the loss function
- Save weights after training!
Autoencoder
- Check representation, plot in 2D:
def plot_representation():
ae,enc = autoencoder()
ae.load_weights('mnist_autoencoder.h5')
encoding = enc.predict(testX)
plt.figure(figsize=(8,8))
for cl in np.unique(testY):
mask = testY == cl
plt.plot(encoding[mask,0],encoding[mask,1],'.',label=str(cl))
plt.legend()
Autoencoder

Autoencoder
- Check reconstruction:
from skimage.io import imsave
def check_images():
ae,enc = autoencoder()
ae.load_weights('mnist_autoencoder.h5')
imgs = ae.predict(testX[:10])
for ix in range(10):
imsave(f'T03_{ix}_original.png',testX[ix])
imsave(f'T03_{ix}_restored.png',imgs[ix])
Autoencoder

Representation Learning
Assignment 1
Assignment 1
Context: image classification
- Super-resolution fluorescence microscopy images of bacteria
- Stage 0: before division starts
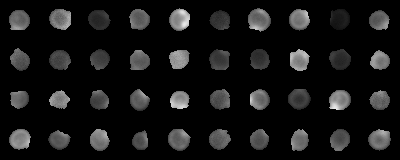
Assignment 1
Context: image classification
- Super-resolution fluorescence microscopy images of bacteria
- Stage 1: septum starts to form
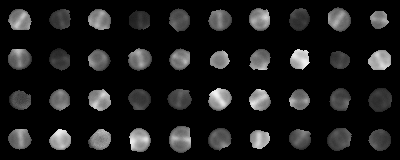
Assignment 1
Context: image classification
- Super-resolution fluorescence microscopy images of bacteria
- Stage 2: septum is formed, before cell splits
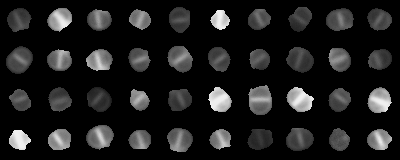
Assignment 1
- Data:400 labelled images, 3892 unlabelled images
- (40x40 pixels, grayscale)
data = np.load('data.npz')
X_u = data['unlabelled'].astype("float32") / 255.0
X_l = data['labelled'].astype("float32") / 255.0
Y_l = keras.utils.to_categorical(data['labels'],3)
- Tasks: experiment, justify and discuss architectures for
- A classifier using the 400 labelled images (300 train, 100 validation)
- An autoencoder with 3892 unlabelled images (use the 400 for validation)
- A simpler classifier using the encoded representations of the 400 labelled images (300 train, 100 validation)
- Similar to the exercises we have done so far.
- Deadline: March 29 (ideally)